- Proteomics Approach for Discovering Renal Cell Carcinoma Biomarkers
- Introduction: Proteomics Approach for Discovering Renal Cell Carcinoma Biomarkers
- Discussion
- The principle of the current proteomic method for biomarker discovery is:
- Use of Proteomic method for discovery and validation of biomarker required for Kidney Cancer: (Application)
- Application of Proteomic in Kidney Cancer:
Proteomics Approach for Discovering Renal Cell Carcinoma Biomarkers
The UK's top-notch assignment writing service, Native Assignment Help, specialises in providing unparalleled quality and outstanding customer satisfaction through our human-written assignment. Our dedicated team can readily assist you with challenging essays or intricate research papers. Choose Native Assignment Help today and experience first-hand what a reliable and committed partner can achieve for you.
Introduction: Proteomics Approach for Discovering Renal Cell Carcinoma Biomarkers
The purpose of this essay is to shed light on “biomarkers”. More specifically it can be stated that this particular essay is aiming to highlight one specific biotechnology method used widely to discover biomarkers and to ensure its validation and implementation to detect any disease. For this proposed essay, the Proteomics approach or method has to be taken into consideration as an instance of a biotechnology method to discover biomarkers for Cancer. For this proposed assessment, focus can be given to Renal Cell carcinoma. More specifically, in this proposed work, the focus will be given on Renal Cell Biomarkers used to treat kidney cancer.
Discussion
According to Azuaje et al., (2019), The term "proteome" refers to the total protein complement which can be encoded by certain genome sequences and encompasses post-translational modification of the protein, and its isoform.
In terms of the representation of evidence about the effectiveness of proteomics in the discovery of biomarkers for RCC, focus can be given to the basic Proteomic techniques. There are several resources which clarify that the use of proteomic methods and biomarker discovery is associated with the extraction and identification of the cells, proteomes and other substances. In the Proteomic method, 2DE or two-dimensional gel electrophoresis has been used to separate the mid-range molecular proteins sequentially based on two discrete features: charge and size of the protein. Afterwards, LC-MS or liquid chromatography technique has been used for the discovery of biomarkers by a proteomic approach (Wu and Fenton, 2018).
The current approach in proteomics is targeted proteomics, where the analysis can enable the validation and verification of the biomarker. This process can be divided into two types: MRM or multiple reaction monitoring and Parallel reaction monitoring or PRM, which is used to ensure the quantitation accuracy at the time of validation and verification process in the discovery of biomarkers.
The principle of the current proteomic method for biomarker discovery is:
MRM is a quantitative proteomic method which used QqQ or Triple Quadrupole pr QTRAP. The targeted precursor ion for the peptide is then selected from the Q1 and diffused to the collision cell or Q2 for disintegration. The specific ion derived from the targeted precursor ion is selected in Q3, which is a low-resolution mass analyzer and cannot transmit ions with the isolation width of 0.7-1.0Da and without long sensitivity (Coca, 2018).
On the other hand, PRM is now a targeted acquisition method that uses HRAM or high-resolution accurate mass analyzer or Orbitraps. The basic principle of biomarker discovery is similar to that of MRM, only in the case of PRM, the Q3 is used to be substituted mass analyzer, this can ensure parallel detection of the protein from the targeted peptide (Daniels et al., 2021).
Use of Proteomic method for discovery and validation of biomarker required for Kidney Cancer: (Application)
AKI requiring dialysis or AKI-D is associated with high mortality in patients with kidney cancer. By using the (slow off-rate modified aptamer scan) SOMA scan proteomic approach, some important biomarkers for kidney cancer have been discovered (Xiao et al., 2019). Those are FGF23 or fibroblast growth factor-23, tPA or Tissue plasminogen activator, and IL-6 as potential mortality-associated biomarkers. The quantitative proteome profiling for the AKI sample by using the SOMAscan assay was used to quantify the (approximately 1305) high, middle and low availability of proteins used for the single-stranded DNA slow off-rate modified aptamer, which has 5 calibrations and 2 quality control sample for every assessment (Vasaikar et al., 2018).
In the case of in-vitro analysis uses 50 µl of serum used to be incubated with immobilised SOMAmers. SOMAmers are used to bind with the cognate protein which is released and hybridized through DNA microarray. Using an Agilent C scanner, the microarrays were scanned. The unprocessed data were treated as previously mentioned. Relative fluorescence units (RFUs) were primarily subjected to hybridization control normalisation to remove variation introduced during the hybridization and scanning processes, median signal normalisation to remove predisposition, and calibration to take into account inter-run variations. After that volcano plot of SOMAscan analysis of 1305 serum protein identifies the change of protein folds with the statistical significance of p>0.05 (Daniels et al., 2021).
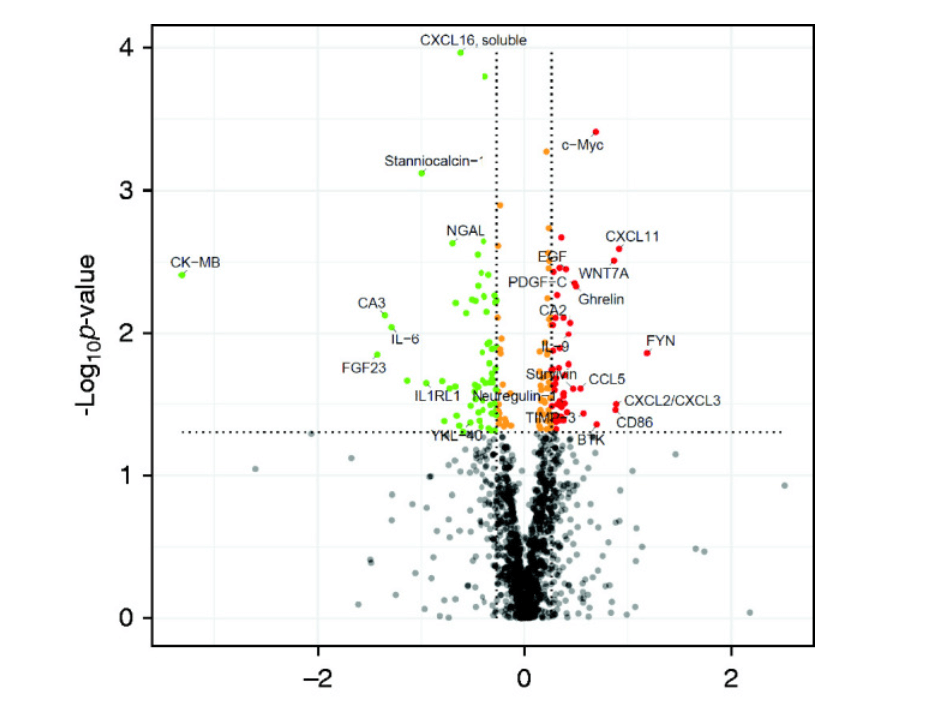
Figure 1: Comparing samples taken on 8th day from individuals with improved kidney and another sample are taken after 28 days incubation from patients whose kidney function had not improved.
In the above figure, the red dots show the fold change with high specificity, and the green dots indicate the protein with low specificity after incubation of the target proteins for 8 days. In the graph, the vertical line in the plot refers to the 1.2 fold of changes in up and down expression and the horizontal line represent the P value of 0.05. the dots in the plot represent the isolation and discovery of the biomarkers for AKI, for example, C-X-C motif chemokine 11 (I-TAC); CXCL2/CXCL3, Tyrosine-protein kinase BTK, metalloproteinase inhibitor 3, creatine kinase-MB, fibroblast growth factor 23, IL-6 and so on which are actively used as a biomarker for the kidney cancer.
The below figure indicates the discovery of novel predictive biomarkers for Kidney recovery for patients with Dialysis -dependent AKI.
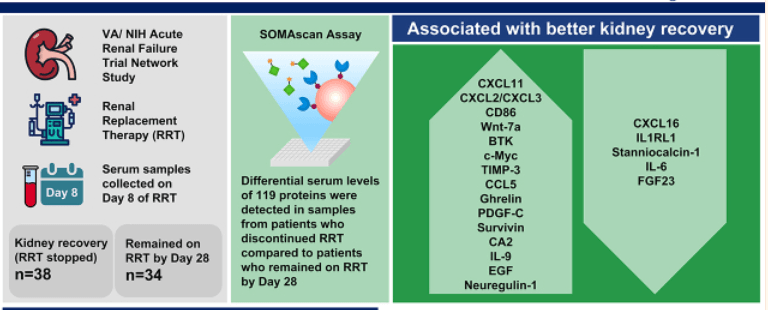
Figure 2: Process of discovering the Renal cancer biomarker by proteomic method.
Application of Proteomic in Kidney Cancer:
A comprehensive understanding of the metastatic pathway is important to treat kidney cancer. By using the proteomic method and analysis, the biochemical confirmation of cleavage of the substrates can be performed and in the case of kidney cancer the cleavage sites can be identified by MALDI-TOF. Kidney cancer or RCC (renal cell carcinoma) is considered the sixth major cause of cancer death in the US and across the world. By using the Proteomic method, the pathway and network approach can be used to identify the biological process associated with the clear cell RCC or ccRCC (Vasaikar et al., 2018). If a patient with renal cell carcinoma is being diagnosed by using SOMAscan (which is an innovative multiplex proteomic platform to measure>1300 protein), 46 spots can be identified with a high degree of confidence.
To confirm the validity of the RCC biomarker discovery focus can be given to the identification of Hsp27 and PKM2 protein. To confirm that the proteomic analysis utilization towards the discovery of the biomarker for RCC is valid indeed, it is important to perform further analysis of two highly unregulated sports : PKM2 and Hsp27 (Vasaikar et al., 2018). The Hsp 27 lies downstream of the p38MAPK, a heat-shock protein that plays an important role in cellular processes like apoptosis. The abundance of Hsp27 can increase the immunoblotting and confirm the proteomic data. In the case of RCC or kidney cancer, the proteomic analysis shows a high degree of phosphorylation of Ksp27 in comparison to normal renal cells. On the other hand, the proteomic analysis ensures the validation of PKM2 as the biomarker for Kidney cancer by immunoblotting (Xiao et al., 2019). In the proteomic assay, it is shown that in absence of pVHL (VHL-deficient RCCs in VHL disease), HIF-1 α used to remain in an active state and the tissue in renal cells acts in a hypoxic way. PMK2 has important in RCC and it can be transcriptionally activated by HIF-1. On the other hand, the hypoxic treatment of the cancer cell line, specifically the renal cell line can cause the increase in PKM2 mRNA which suggests that this protein biomarker is important and valid in VHL-deficient RCCs. And these results confirm the validation and verification of the importance of PMK2 as a biomarker which increased in amount in the tumour tissue examination.
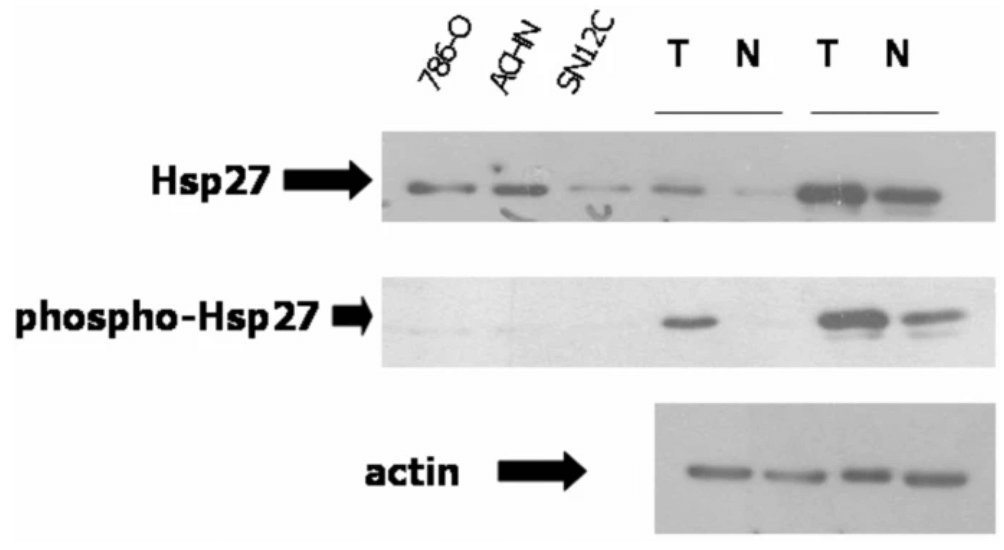
Figure 3: Anti-apoptotic protein is in the unregulated condition in ccRCC or kidney cancer, confirmed by the Proteomic analysis and immunoblotting. Three RCC cell lines and two tumours are used in the proteomic analysis and immunoblotted with Hsp27 antibodies, actin has been loaded as control.
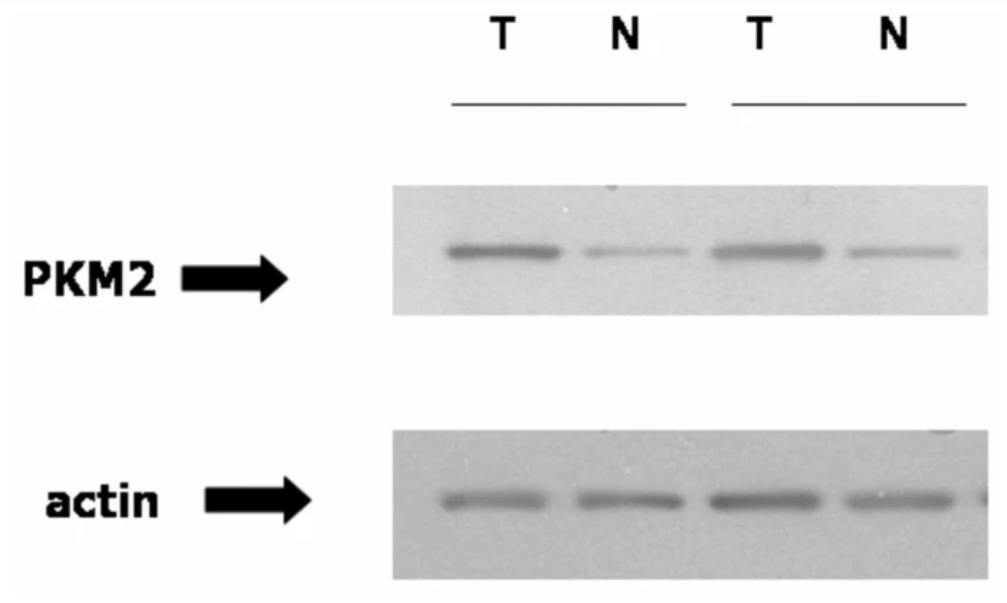
Figure 4: PMK2 is increased in ccRCC which is confirmed by the immunoblotting and ensures the validation and verification of Kidney Cancer biomarkers by Proteomic method. In the Immublotting process, PMK2 antibody is used and actin is loaded as control. The solid line indicates the same kidney.
In the Proteomic method,sensitivity of protein can be measured by compiling the protein copy number. the copy number distribution can be done by the Q-Exactive HF system or chromatography system on the Q-Exactive classic process of protein counts.
Conclusion and Limitation
To conclude this essay, it can be stated that in current times, the importance of Proteomics technologies in the discovery and validation of biomarkers for any disease can never be denied. Though considering the entire essay, it can be stated that there are several limitations related to proteomic technology in the discovery of cancer biomarkers. The major disadvantage is, the proteomic technology depends on the protein and peptide patterns, and characteristics of post-translational modification, though, currently, these technologies are under development and the process is lack standardization in sample processing.
As per the estimation of Proteomics shared resources, this technology used to cost $80 per hour (Orbitrap Fusion) on the mass spectrometer to develop Cancer biomarkers. Identification of the protein by using immunoblotting or SOMA scan costs approximately $400. This is one of the core challenges related to the use of Proteomic technology for biomarkers discovery and managing the validation. Though, Proteomic technology is now becoming an important approach in biotechnology and molecular biology, as it offers reliable information about the identification, expression, and modification of proteins.
References
Azuaje, F., Kim, S.Y., Perez Hernandez, D. and Dittmar, G., (2019). Connecting histopathology imaging and proteomics in kidney cancer through machine learning. Journal of clinical medicine, 8(10), p.1535.https://www.mdpi.com/540750
Coca, S.G., (2018). “Scanning” into the Future: The Promise of SOMAScan Technology for Kidney Disease. Kidney International Reports, 3(5), p.1020.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6127447/
Daniels, J.R., Ma, J.Z., Cao, Z., Beger, R.D., Sun, J., Schnackenberg, L., Pence, L., Choudhury, D., Palevsky, P.M., Portilla, D. and Yu, L.R., (2021). Discovery of novel proteomic biomarkers for the prediction of kidney recovery from dialysis-dependent AKI patients.Kidney360,2(11), p.1716.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8670726/
Goossens, N., Nakagawa, S., Sun, X. and Hoshida, Y., (2015). Cancer biomarker discovery and validation.Translational cancer research,4(3), p.256.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4511498/
Li, D. and Chan, D.W., (2014). Proteomic cancer biomarkers from discovery to approval: it’s worth the effort.Expert review of proteomics,11(2), pp.135-136.https://www.tandfonline.com/doi/abs/10.1586/14789450.2014.897614
Percy, A.J., Chambers, A.G., Smith, D.S. and Borchers, C.H., (2013). Standardized protocols for quality control of MRM-based plasma proteomic workflows.Journal of proteome research,12(1), pp.222-233.https://pubs.acs.org/doi/abs/10.1021/pr300893w
Sallam, R.M., (2015). Proteomics in cancer biomarkers discovery: challenges and applications.Disease markers,2015.https://www.hindawi.com/journals/DM/2015/321370/
Vasaikar, S.V., Straub, P., Wang, J. and Zhang, B., (2018). LinkedOmics: analyzing multi-omics data within and across 32 cancer types.Nucleic acids research,46(D1), pp.D956-D963.https://academic.oup.com/nar/article-abstract/46/D1/D956/4607804
Wu, Q. and Fenton, R.A., (2018). Proteomic approaches in kidney disease biomarker discovery. American Journal of Physiology-Renal Physiology, 315(6), pp.F1817-F1821.https://journals.physiology.org/doi/abs/10.1152/ajprenal.00421.2018
Xiao, Y., Ma, D., Zhao, S., Suo, C., Shi, J., Xue, M.Z., Ruan, M., Wang, H., Zhao, J., Li, Q. and Wang, P., (2019). Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer.Clinical cancer research,25(16), pp.5002-5014.https://aacrjournals.org/clincancerres/article-abstract/25/16/5002/125167